The Rebirth of Manufacturing?
In today’s aggressive, cost-cutting climate, the desire to switch from batch to continuous manufacturing is strong. As pharma companies embark on prolonged innovation cycles in an attempt to deliver the higher volumes and reduced costs of continuous manufacturing, it might surprise many that the technology they seek already exists.
“This is the book for starting the next century of engineering.” High praise indeed, especially when it comes from Marvin Minsky (

The factory in a box
Drexler describes molecular manufacturing as the process of continuously bonding molecules together to produce larger structures, ultimately assembling components whose sizes and complexities range between that of a spatula and an airplane. Bonding is controlled and directed by so-called molecular assemblers, which resemble the machines found in factories today. Drexler packages this vision as ‘3D-printing perfected’ – nanofabrication engines that are quintessential factories-in-boxes (
There is no doubt that Drexler is a visionary and a consummate author. However, when scientific ideas run the risk of being confused for science fiction (Drexler himself raises the example of the Star Trek replicator (
To understand Smalley’s criticisms, it is helpful to visualize the molecular assembler as a collection of flexible appendages. Since the appendages themselves are also composed of molecules, it is inevitable that the size of these constituents may exceed that of the molecules being fused together – a scenario that becomes particularly relevant at the start of an assembly cascade. As a consequence, the appendages might not be dexterous enough for atomically or molecularly precise fabrication, much in the same way a person with fat fingers might struggle to type long sentences on smartphones with small screens. Atomic-level attractive forces between the appendages and the substrate molecules could also interfere with the fabrication process – a situation that is akin to making origami with gluey fingers. What a mess!
Needless to say, Drexler did not take too kindly to this criticism, and, in a strongly worded rebuttal (
While the Nobel Laureate did concede that a machine such as a ribosome is indeed capable of precise fabrication, he raised doubts regarding the capacity of Drexler’s nanobots to exhibit the same degree of control and self-repair as a ribosome. Smalley was also unconvinced about the ability of biological systems to perform non-aqueous chemistries or synthesize inorganic species, such as silicon, or produce materials of construction, such as steel and concrete.
Smalley does not seem to have been an avid follower of developments in the life sciences. In any case, he was certainly oblivious to biomineralization (
Nevertheless, of all the surprises thrown up by the debate, the one that stood out the most was the low opinion that Smalley and Drexler shared of biological manufacturing platforms. Even after all these years, Drexler’s views have not changed in the slightest. Arthur C. Clarke – one of the greatest science fiction writers – said it best: “If an elderly but distinguished scientist says that something is possible, he is almost certainly right; but if he says that it is impossible, he is very probably wrong.”
Metabolic engineering
At the outset, let us re-imagine the cell as a highly connected and well-regulated network composed of a large number of chemical reactions, wherein atoms are sequentially added, removed or exchanged from a molecule as it proceeds through the network. Each reaction is catalyzed by a unique enzyme, which itself is a product of a specific gene. A metabolic network, therefore, is a biological analogue of Drexler’s molecular assembly line – a true microfactory. Each enzyme retains its role of a biological machine, and the genetic make-up of the cell is equivalent to the blueprint of the factory floor. Similarly, a metabolic pathway is simply a sequence of interacting machines.
Now, if one were to express an additional copy of a specific gene – altering the factory’s blueprint very slightly – the presence of duplicate machines executing the same task would increase the productivity of that particular step. Since no two biochemical reactions are the same, increasing the productivity of one machine in the step could have several possible outcomes. The change could serve to increase the productivity of subsequent machines in the assembly line if these machines are operating below their maximum capacities, meaning that the rate of movement of material through the pathway – a quantity that is termed as ‘flux’ – increases as a whole. In the converse scenario, if the downstream machines are already functioning at peak capacity, the stock of product produced by the machine in question will begin to grow.
Regardless of the outcome, owing to the high degree of connectivity within the network, manipulation of a single step in the network will spawn system-wide perturbations that greatly alter the intracellular concentrations of several metabolites. Similar effects will also be observed when genes are deleted or manipulated by modifying their sequences, or when foreign genes are introduced into the genetic blueprint. In the latter two scenarios, the genes will encode novel enzymes that synthesize altogether new products by siphoning away atoms destined for native metabolites.
Altering the genetic make-up of a cell to achieve heightened or entirely new production of a molecule is defined as metabolic engineering, and operational decisions such as whether a gene should be deleted or overexpressed – and if it is overexpressed, by how much – are governed by the stoichiometry, kinetics and regulation of the pathway of interest. Therefore, metabolic engineering combines deep understanding of metabolism, biochemistry and molecular biology with the rigorous quantitation of fluxes (
To his credit, Drexler did agree that ribosomes and enzymes are extremely efficient molecular assemblers. He just wasn’t sure if these biological machines could ever be employed for synthesizing usable materials beyond common biochemicals, let alone suggest how they could be deployed for next-generation, sustainable manufacturing. Fortunately for us, recent innovations in analytical chemistry, genome sequencing and assembly, and bioinformatics, as well as quantum improvements in the tools of molecular biology have not only brought to light the vastness and staggering diversity of nature’s biocatalytic repertoire (
Greener pharma
The prospects in store for the trillion-dollar pharmaceutical industry are particularly exciting. The vast majority of manufacturing processes in the pharmaceutical industry comprise energy- and resource-inefficient batch operations, and these processes generate, on average, anywhere between 25 and 100 kilograms of waste for every kilogram of product (
Examples of pharmaceutical companies closing down manufacturing operations because waste disposal costs are overtaking the selling price of the product are becoming increasingly common (for example, DSM shut down manufacture of phloroglucinol, a therapeutic agent used to treat gastrointestinal disorders). There is a clear and compelling opportunity to improve the efficiency of manufacturing processes by transitioning from batch to continuous operation.
In light of some of the aforementioned scientific and technological developments, metabolic engineering is arguably best placed to deliver the revolution in continuous API manufacturing that the industry is so desperately seeking. As the pharmaceutical industry currently spends
Sustainable Scale Up
Professor Bernhard Palsson is CEO of the Novo Nordisk Foundation Center for Biosustainability at the Technical University of Denmark, which aims to solve some of the scientific and engineering challenges of large-scale metabolic engineering. We caught up with Palsson to find out more.
What is the goal of the Center?
The Center is the first large-scale research operation that is solely focused on the design, construction and testing of cell factories. The creation of microbial production strains is often very expensive and time consuming – around $50 million and five years. Our goal is to cut those figures to less than $20 million and two years. To do that, the whole center is organized around an iterative design workflow, so that we focus our resources on the current rate-limiting step in the process.
What are some of the most exciting ongoing projects?
There are a couple of pathways that generate compounds important to human physiology, like serine, adrenaline, L-Dopa, melanin, serotonin, nicotine and caffeine. The Center has an exciting project underway to build these pathways into E. coli or yeast, so that we can make any of these compounds in any quantity we like, at an acceptable price.
In the field of protein engineering, one of our scientific directors, Henrik Clausen, has mapped out an engineering platform for specifying glycans on IgG, produced in CHO cells. By customizing glycosylation in the CHO cells that are used to produce IgG, there is potential to improve product homogeneity and therapeutic efficacy.
Where do you think the field is heading?
Taken together, biosustainability efforts for these compounds could have a huge impact on the world’s economy – five percent of the world’s GDP comes from chemical or pharmaceutical manufacturing. At the Center, we have been trying to determine how many of the tens of thousands of commercially available chemicals can be made biologically, and it appears that it is a very sizeable fraction. The combination of basic research into metabolic pathways and host cell optimization for effective scale up is likely to make enormous inroads into the manufacture of biochemicals and pharmaceuticals over the next decade or two.
roughly a quarter of its annual revenues on manufacturing (
And there’s another advantage of using metabolic engineering for production of pharmaceuticals – even the quality and safety of drugs could vastly improve (
A typical pharmaceutical manufacturing process can be divided into two distinct stages – API synthesis and drug formulation. Purified API is produced in the first stage via an iterative sequence of reaction, separation and purification steps. The API is then suitably formulated during the second stage to yield the finished dosage form. Since nearly 80 percent of pharmaceutical products in the market today are tablets, the product from the first stage is often in a dry, crystalline form. Tablet formulation commences with milling of the API crystals to form a fine powder, which is then blended with excipients and mixed with water to produce wet granules. Drying, tableting and coating follow suit, and the manufacturing process culminates with packaging of the tablets for distribution and sale.
Of all the steps listed above, only milling and tableting currently happen continuously. Clearly, the scope for improvements in the pharmaceutical manufacturing chain is immense. Innovations have indeed occurred, notably in processes that comprise the second stage. For instance, the use of continuous crystallizers, mixers and blenders, and granulators are slowly taking root, and Novartis recently demonstrated continuous operation of the entire second stage of the manufacturing process for Diovan, a drug used to treat high blood pressure and heart disease, at the pilot scale (
On the synthesis front, however, the situation is less encouraging. Seventy percent of all reactions that are employed for synthesizing APIs are still undertaken in batch mode (
The solution?
API synthesis is an iterative process involving multiple reactors and separation equipment. Briefly, one or more raw materials are fed to the first reactor in the sequence along with the requisite reagents, solvents, catalyst and energy. Following completion of the reaction, the ‘product’ – namely, the first intermediate in the synthesis scheme – is purified from the reaction mixture using one or more separation processes, such as filtration or solvent extraction. More often than not, though, the solvents and reagents that are employed in these steps are noxious substances, and large quantities of these materials – not to mention energy – are consumed before a small amount of the intermediate is produced. The first intermediate is then mixed with a different set of reagents, solvents and catalyst in the second reactor, where it is eventually converted to the second intermediate. This sequence of reaction–separation steps continues until the final API is synthesized. Then, the API is crystallized and dried, and later sent to the formulation unit for production of the finished doses.
Now, in the very same process train, imagine a situation wherein no harmful solvents or reagents are ever consumed, and atoms in the raw material supplied to the first reactor are precisely rearranged to produce the first intermediate without ever requiring the temperature of the reaction mixture to rise beyond ambient conditions. The one-to-one conversion of raw material to the first intermediate also obviates the need for any purification. Next, imagine that the first intermediate shuttles over to the next reactor without any expenditure of energy and then undergoes yet another precise, reagent-free rearrangement to form the next intermediate. This sequence of ‘clean transformations’ continues until the API is synthesized. Sound familiar?
In nature, cheap and abundant carbohydrate-based raw materials are continuously transformed to APIs within a single cell, and what was once an extended cascade of reaction and separation steps is done away with and replaced by a configuration comprising a single reaction–separation step. The synthesis of taxadiene, an intermediate in the taxol biosynthetic pathway, perfectly encapsulates the jaw-dropping efficiency possible. The most efficient chemical synthesis of taxadiene reported to date involves 18 reaction–separation steps (
The API itself can be extracted from the culture broth using continuous countercurrent chromatography (
The big question
What is the range of products that can be manufactured? Well, this is where metabolic engineering arguably provides its greatest value; the success rate of a conventionally synthesized drug candidate is one in 100,000; on the other hand, one in every 350 natural products that are screened for drug activity eventually makes its way to market. There’s a good reason for the nearly 300-fold difference between the success rates of synthetic chemicals and natural products – the latter typically have more chiral centers, rings, oxygenated substituents, and solvated hydrogen-bond donors and acceptors, which reduces the entropic costs that these molecules incur during binding to drug targets. Unsurprisingly, natural products make up well over half of the modern pharmacopoeia, despite the industry tapping into only a small fraction of this roughly 170,000-strong pharmaceutical treasure chest (
However, with the advent of metabolic engineering, the equation is finally shifting back in favor of natural products. Recent investigations into natural product biosyntheses have revealed that these complex molecules are assembled programmatically by pathways that are combinations, permutations and mutations of only a
Pioneering Pathways
Researchers around the world are working to find more sustainable ways of producing plant-derived drugs. We spoke to Jay Keasling, Professor at University of California, Berkley, about his pioneering work on the malaria drug artemisinin, which has been described as the “poster child for synthetic biology”.
What makes artemisinin a good target for metabolic engineering?
It’s a molecule that has already been approved for the treatment of malaria and is widely used. It’s derived from the Wormwood plant, which is not difficult to grow, but the production times are long and it has been difficult for companies to predict demand. Coupled with unpredictable crop success, it’s been a challenging drug to source and a lot of it is needed. Finally, we had been working on chemicals that were in the same compound family as artemisinin, so we knew it should be possible to make it.
How did you go about synthesizing the drug?
Several of the genes encoding the metabolic pathway for artemisinin were already known, but some were not; the first job was studying the plant to find those enzymes. That was challenging because plants have a huge number of genes. To find the single gene responsible from 50,000 is a big deal – especially 10 years ago when we started – we didn’t have the high throughput sequencing technologies we have now. Once we had the pathway, we tried it out in both E. coli and yeast, and found yeast to be the most effective. The first 1.7 million treatments made using this method were shipped in August. It feels great when something you have worked on benefits people.
Where has your research taken you since then?
The power of metabolic engineering is that once you have built a platform like the one for artemisinin, then it’s not so difficult to make a few changes to the pathway and make related compounds. It turns out that the same compound family as artemisinin also includes molecules that can make diesel, jet fuel and even fragrances.
We are also continuing our work on pharmaceutical applications. There’s a molecule called prostratin, derived by healers in Samoa from the bark of the mamala tree, and used to treat viral infections. Ethnobotanist Paul Cox came across prostratin while in Samoa looking for an anticancer drug, and it was found to have possible applications for HIV therapy. One of the big problems with HIV is that you can kill the active virus, but not the latent phase. By activating the virus, prostratin could allow antiretroviral therapies to eliminate that reservoir. The drug is now approaching clinical trials, and we are working on determining the metabolic pathway involved, to find a reliable production method.
There are drugs that have been abandoned at clinical trials because the drug simply couldn’t be sourced in sufficient quantities from a natural source. Metabolic engineering gives us a method to produce drugs that we otherwise couldn’t.
handful of enzymatic reactions. In other words, altering the order, number and cross-talk between the biological machines in the cellular microfactory, or minutely tweaking them, could provide access to a staggering collection of natural products, instantly transforming metabolic engineering into a tool for drug discovery. There are other benefits too, such as ease of scale-up. In order to go from gram-scales to ton-scales, one simply has to use a larger fermenter! The footprint of a manufacturing facility based on metabolic engineering is also considerably smaller.
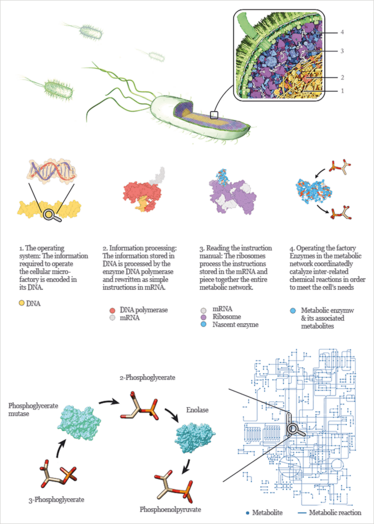
Figure 1. A glimpse into the inner workings of a bacterial cell.
The big challenges
So why isn’t metabolic engineering in widespread use in the pharma industry? There are two stumbling blocks. The first is a general lack of good tools. The tools used for manipulating a microbial host’s metabolism are not universally applicable. Indeed, they are specific to only certain pathways or products. The lack of transferability of the majority of tools and techniques between hosts, in turn, can be attributed to an incomplete understanding of the regulatory mechanisms in the cell (more on that below). Clearly, there is an acute need for transferable tools and techniques that can characterize and manipulate host regulatory mechanisms to control heterologous over-production of natural products and other secondary metabolites.
Second, there are some significant technical and knowledge roadblocks. Because most natural products are components of the immune and defense systems of their host – and because an organism’s evolutionary fitness is contingent on its ability to synthesize that otherwise rare molecule – the vast majority of biosynthetic pathways comprise enzymes that act on several substrates and/or catalyze the formation of multiple products. And that implies that the pathways are actually highly branched, possibly synthesizing several products. The paclitaxel biosynthetic pathway, for example, generates well in excess of a hundred products. As natural product biosynthetic pathways are quite long, the dissipation of intermediate molecules to competing chemical reactions at each step amounts to inordinate losses in the overall yields. Enhancing the substrate and product fidelities of dissipative enzymes is now an urgent problem in metabolic engineering and biocatalysis. Unfortunately, the established method for modulating the activity of an enzyme (which involves selecting similar examples in nature as starting points for modification using mutagenesis or directed evolution) is not applicable to the current scenario. More elaborate structure-guided approaches are required. However, not only is this a very long and involved process, but even in the event that the characteristics of every single enzyme in the pathway are eventually improved, it is uncertain whether simple microorganisms, such as E. coli, will be able to express such a large collection of enzymes without grave physiological stress.
Diversity-oriented biosynthesis presents an entirely new challenge for metabolic engineers, and it is apparent that synthesizing an advanced intermediate that acts as a gateway molecule for target-oriented chemical synthesis is a more viable alternative. Not only would the number of enzymes be significantly more tenable, but such a semi-synthetic manufacturing process would also take advantage of the core competencies of both metabolic engineering and synthetic chemistry.
Despite the hurdles, progress has been rapid in recent years, with groups all around the world making huge strides (see sidebars, “Sustainable Scale Up” and “Pioneering Pathways”).
Metabolic engineering truly is the ultimate demonstration of continuous manufacturing and is well on its way to evolving into a very handy tool for drug discovery. These developments are the stuff of Drexler’s wildest dreams. Perhaps now he might reconsider his stance regarding biology.
- G. L. Challis and J. H. Naismith, Curr. Opin. Struct. Biol. 14, 748-756 (2004).
- K. E. Drexler, Wiley, New York, NY (1992).
- K. E. Drexler, The Guardian, bit.ly/1ohlk8i (14 October 2013).
- K. E. Drexler, The Guardian, bit.ly/1eblzqw (21 October 2013)
- R. E. Smalley, Scientific American 285 (3), 76-77 (2001).
- K. E. Drexler et al., Institute For Molecular Manufacturing, bit.ly/1t3w80j (2001).
- P. P. Dennis and H. Bremer, J. Molec. Biol. 84, 407-422 (1974).
- R. Baum, Chemical & Engineering News 81 (48), 37-42 (2003).
- S. Mann, Nature 365, 499-505 (1993).
- G. Stephanopoulos, Metab. Eng. 1 (1), 1-11 (1999).
- L. P. Wackett and C. D. Hershberger, American Society Microbiology, Washington, DC (2001).
- M. T. Reetz, J. Am. Chem. Soc., 135 (34), 12480-12496 (2013).
- U. T. Bornscheuer et al., Nature, 485, 185-194 (2012).
- C. Khosla and J. D. Keasling, Nat. Rev. Drug Discov. 2 (12), 1019-1025 (2003).
- V. G. Yadav et al., Metab. Eng. 14 (3), 233-241 (2012).
- R. A. Sheldon, Green Chem. 9, 1273-1283 (2007).
- D. Weiss, P. Naik and R. Weiss, Nat. Rev. Drug Discov. 8, 533-534 (2009).
- K. Plumb, Chem. Eng. Res. Des. 83 (A6), 730-738 (2005).
- G. S. Calabrese, S. Pissavini, Alche J. 57 (4), 828-834 (2011).
- G. Malhotra, Pharm. Processing 27 (8), 16-20 (2010).
- V. G. Yadav, Ind. Eng. Chem. Res., 53 (49), 18597–18610 (2014).
- S. Y. Rojahn, MIT Technology Review, bit.ly/1keso2b (10 May 2012).
- D. M. Roberge et al., Chem. Eng. Tech. 28 (3), 318-323 (2005).
- Pennemann, H. et al., Org. Process Res. Dev. 8, 422-439 (2004).
- S. M. Rubenstein and R. M. Williams, J. Org. Chem. 60, 7215-7223 (1995).
- V. G. Yadav, Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA (2013).
- A. Jungbauer and J. Peng, Biotechnol. J. 6, 1431-1434 (2011).
- B. M. Trost, Science 254, 1471-1477 (1991).
- R. Firn, Oxford University Press, Oxford, UK (2010).
As an undergraduate student in chemical engineering at the University of Waterloo, Vikramaditya G. Yadav coveted a career in Alberta’s burgeoning petrochemical sector. One evening during his second year, he stumbled upon a copy of Juan Enríquez’s ‘As the Future Catches You’ in the library and was instantly captivated by biological engineering. “My new found passion took me on a wonderful journey that included stops at Sanofi Pasteur and the Massachusetts Institute of Technology, where I received my doctoral degree for a thesis on engineering enzymes and bacteria for synthesis of pharmaceuticals,” says Vikramaditya. Now, as an Assistant Professor at the University of British Columbia, he is conducting wide-ranging research in metagenomics, bioenergy, water bioremediation, drug discovery and manufacturing, phytochemistry and Alzheimer’s disease.



















